Gene Expression Regulation pre-mRNA Processing
5'-end capping
5'-Capping occurs shortly after transcription initiation, when nascent transcripts are 20-25 nt in length. The Pol II CTP becomes hyperphosphorylated during the transition into productive elongation. The unphosphorylated CTD exists closely associated with Pol II core, as well as associated factors such as TFIIE. Phosphorylation dissociates the CTD so that it tails out behind the Pol II elongation complex. The end of the CTD tail binds two enzymes, capping enzyme (CE), and N7G methyltransferase (MT) (Shatkin and Manley, 2000).
Mammalian CE catalyzes two reactions:
RNA-5'-triphosphatase
pppN(pN)25 ppN(pN)25 + Pi;
guanylytransferase
ppN(pN)25 + GTP GpppN(pN)25 + PPi;
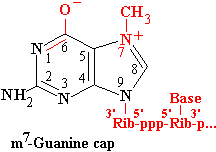
The mammalian CE is a single bifunctional enzyme wherease yeast CE exists as two separate subunits. Appropriate fragments of the mammalian enzyme can complement defects in the respective yeast subunits. G becomes linked to the triphosphate through its 5'-O, so has the opposite orientation to the other nucleotides in the chain.
MT methylates the attached G on N7. Methylation in this position gives rise to a quarternary N+, changing G into its enolate form. In many cases, the initiating nucleotide is A, which can be N6 methylated as well.
methyltransferase
GpppN(pN)25
+
S-adenosylmethionine m7G-ppp-N(pN)25
+
S-adenosylhomocysteine;
Capping protects the nascent pre-mRNA against degradation; failure to cap or loss of cap leads to rapid breakdown of the RNA. The m7G-cap is bound by two nuclear Cap Binding Proteins, CBP20 and CBP80. Only CBP20 appears to be in direct contact with the cap. These proteins play a role in initiating subsequent splicing reactions, and anti-CBP antibodies inhibit subsequent splicing reactions. Overexpression of short capped transcripts with no splicing sites can also inhibit splicing by competing for splicing factors.
RNA Splicing
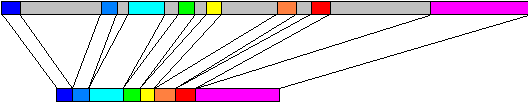
Metazoan genes commonly contain intervening sequences inserted into the open reading frame. The insertions are called introns (gray in the figure above) while the expressed fragments are called exons (coloured in the figure above). Introns must be removed by RNA splicing, which occurs concurrently with transcription. The long upper box in the figure represents the genomic DNA of ovalbumin, 7700 bp, mapped onto the mRNA, about 1900 nt, shown in the lower box. Introns occur less frequently and are much smaller in lower eukaryotes such as yeast.
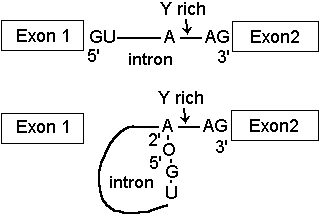
Removal of introns from pre-mRNA transcripts involves cleavage at the 5'- end of the intron by attack of a specific 2'OH group, the branch site. This forms a phosphodiester bond with the 5'-phosphate of the intron, creating a lariat structure.
The intron lariat is then removed, proceeding by attack of 3'-OH on Exon 1 to displace the intron from the 5'-phosphate of Exon 2.
During the whole process, the number of phosphodiester bonds remains constant, so this is not an endonuclease cleavage and ligation process as occurs in tRNA processing, but an ATP independent transesterification.
At step 1, the phosphodiester bond between Exon1 and intron is converted into the 2'-branch site phosphodiester.
At step 2, the phosphodiester bond between intron and Exon 2 is converted into the Exon 1 - Exon 2 phosphodiester.
Some single-celled eukaryotes, e.g. the cilate Tetrahymena, produce pre-mRNA with self splicing introns. In these cases the intron forms a unique tertiary structure promoting self catalysis. The catalytic action is embodied in the RNA itself.. In some examples, catalysis involves attack by the 3'-O of a separate bound molecule guanosine nucleotide, and in other cases the 2'-O of an in-chain A produces the lariat structure.
Autocatalysis is mediated via a metal ion, Mg2+ or Ca2+, bound to a specific site formed by the tertiary structure of the intron. Self splicing introns have been used as the models for ribozymes, or catalytic RNA. For additional information, see Ribosomes and Ribozymes
In most eukaryotes, splicing is mediated by a large ribonucleoprotein complex comparable in size to the ribosome or polymerase II holoenzyme called the spliceosome.
The spliceosome contains a specific set of U-rich small nuclear ribonucleoproteins or snRNPs.
Sequence of reactions in the splicing process
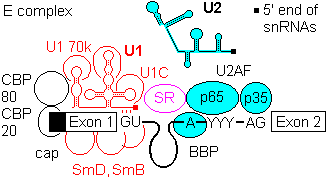
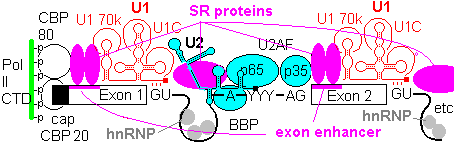
1a) Formation of the commitment or E complex involves binding of factor U1 snRNP (complex of U1 RNA, U1A RRM protein, U1C and U1 70K protein) to the 5'-intron GU site. Recognition is by base pairing of the 3' end of U1 with the consensus sequence AG|GUAGGU (vertical bar is the exon-intron junction), and ATP is consumed in the base pairing process.
SR accessory factors associate with the exon towards the 5' direction, and facilitate binding of U1.
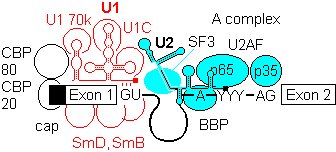
1b) This is generally followed by binding of U2 auxilary factor U2AF (Mud2 in yeast) to the pyrimidine rich tract between the branch site and the 3'- end of the intron. U2, plus the associated SF3a/b can then base pair with the metazoan branch point sequence YNYURAY to give the A complex. An additional protein factor, BBP (branch binding protein) binds in the region of the branchpoint A. In yeast, the branch point is more conserved, UACUAAC.
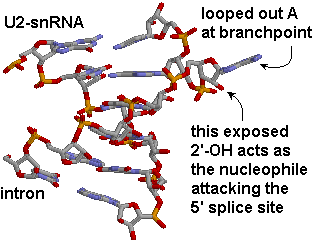
The branchpoint sequence (BPS) is identified by base pairing with a section of the U2 snRNA bearing the sequence 5'-GUAGUA-3'. The BPS sequence is mismatched at a single A, which becomes looped out, exposing its 2'OH. This exposed ribose OH acts as the nucleophile attacking the 5'-splice site.
When the sequence at the branchpoint deviates from the consensus, associated protein factors such as U2AF are needed to promote complex formation.
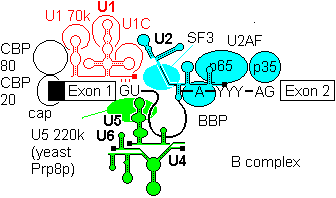
2a) The U4-U6, U5 tri snRNP is then recruited to give the B complex. There is some evidence that U4/U6 recruitment to the 5'-splice site can precede U2 assembly at the branchpoint.
2b) Finally some radical ATP-dependent base pair rearrangements occur to organize the catalytically competent C complex. Two tri-snRNP factors,U5 100p and U5200p have been shown to contain DExD/H box domains.
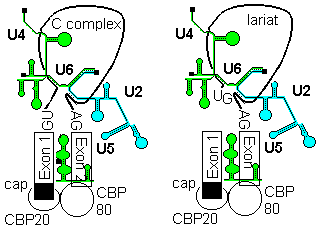
U5 first base pairs to the upstream exon and the 5'-splice site, a process that requires RNA unwindase activity to displace U1 from the exon.
U6 base-pairs to U2, resulting in displacemant of U4. Finally, U5 base pairs to exon 2 near the 3'- splice site on the same stem loop that already holds Exon1, bringing the 3'-OH of Exon 1 into close proximity to 5'-p of exon 2.

Fidelity in splice site determination.
Yeast splice sites tend to adhere closely to consensus sequences, and introns are few and small, so that the basic spliceosomal machinery can recognize sites effectively in most cases. In metazoans, there is more diversity in the splice site environment, introns are large, and multiple splicing of a single pre-mRNA is common. In these conditions, accessory factors are of greater importance to ensure correct splicing.
Exons contain elements called exonic enhancers which are targets for binding SR and related RRM containing proteins. The organization that lays out the splicing pattern starts with the Cap binding complex of CBP20 and CBP80, and possibly even with the CTD of the RNA Pol II . An array of protein factors, e.g. SC35, bind in a cooperative manner between cap and first splice site to define its location. Other SR proteins bridge the intron gapfrom U1 70k to facilitate U2AF binding, and establish branch point and 3' splice site. Once the U2 complex is in place, SR proteins link up to the next 5' splice site, to continue the process. Thus the pattern of splice sites is established progressively from the 5' cap towards the 3' end, and the spliceosome does not select intron targets for splicing at random.
In metazoans, certain members of the hnRNP (heterogeneous nuclear ribonucleotprotein) class bind to sites in particular in the introns. These include hnRNP A1, which binds indiscriminately to pre-mRNA and has a negative effect on spliceosome assembly. The function of the enhancers and SR proteins seems to be to exclude hnRNA from the exons, and a gap in the chain of enhancers and SR proteins allows hnRNP A1 to act as a splicing repressor.
Splice site specificity is reasonably conserved across species, allowing expression of transgenes. Occasionally splice sites may be misread, for example when wild type Green Fluorescent Protein is expressed in higher plants, the polypeptide may be disrupted by misinterpretation of a coding sequence as a plant specific splicing site.