Sorting out the complexity
of SR protein functions
Serine/arginine-rich (SR) proteins play multiple roles in the pre-mRNA splicing process. These proteins are essential for removing introns that are constitutively spliced, as well as regulating alternative splicing. In living cells, SR proteins move from speckles—specialized regions in the nucleus that store splicing factors—to sites where transcription is active. Some SR proteins are also known to shuttle in and out of the nucleus. The localization of SR proteins can be regulated by phosphorylation, which plays a key role in controlling splicing events.
Once inside the nucleus, SR proteins interact with nascent pre-mRNA. While their role in recruiting splicing factors and recognizing splice sites is understood, the precise mechanisms by which they contribute to these processes are still being explored.
SR proteins have a modular structure: an RNA-binding domain at the N-terminus that interacts with pre-mRNA and an RS (arginine/serine-rich) domain at the C-terminus that is involved in protein interactions. It is believed that SR proteins bind to the pre-mRNA and help recruit various splicing factors during the assembly of the spliceosome. They are also thought to mediate interactions between splicing factors bound to the 5' and 3' splice sites.
Recent studies suggest that SR proteins perform all these tasks during each round of splicing, and in some cases, independent SR proteins carry out these functions. This review focuses on the biochemical mechanisms behind SR protein functions in splicing, without delving into their subcellular localization, although this also plays a role in regulating splicing.
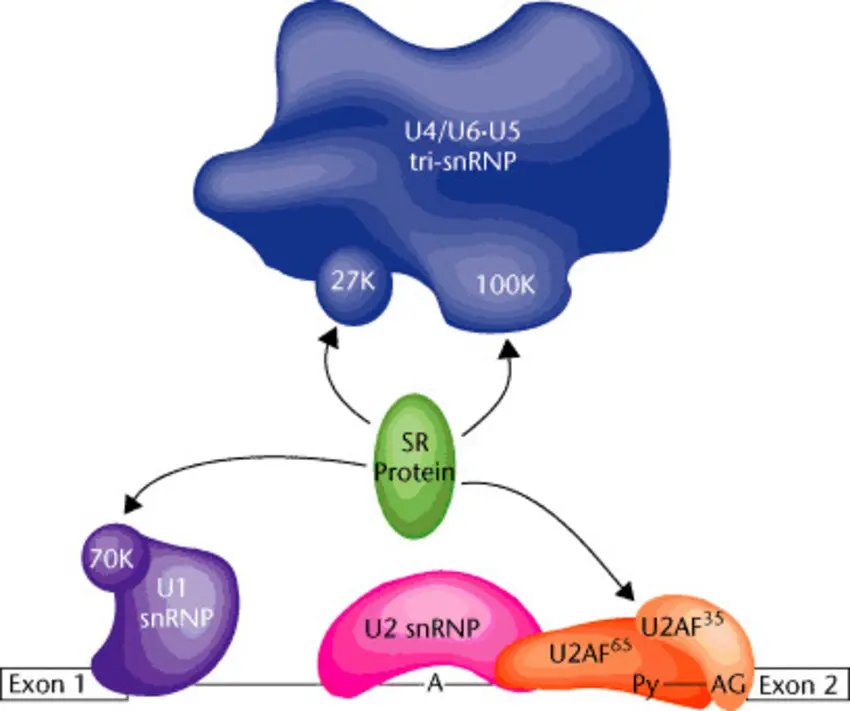
Spliceosome Assembly
The spliceosome is a complex of small nuclear ribonucleoproteins (snRNPs) and non-snRNP proteins that is responsible for splicing pre-mRNA. Its assembly begins when the U1 snRNP binds to the 5' splice site, SF1/mBBP binds to the branchpoint, and U2AF binds to the polypyrimidine tract and 3' splice site. This forms the E complex, a precursor to the active spliceosome.
The assembly continues in a stepwise fashion, with the U2 snRNP binding to the branchpoint to form the A complex. The U4/U6•U5 tri-snRNP then associates to form the B complex, which eventually rearranges into the catalytically active C complex. SR proteins assist in the early stages of spliceosome assembly, including the recruitment of U1 snRNP and U2AF to the splice sites. They may also function in later stages, such as during the transition from the A to B complex.
The SR Protein Family of Splicing Factors
SR proteins were first identified in various studies using different methods. For example, SF2/ASF was purified from HeLa cell extracts as a factor needed to reconstitute splicing. This led to the discovery of other SR proteins, including SC35. These proteins are characterized by their high serine and arginine content, and the family now includes several known members in humans.
The SR protein family is essential for proper splicing in metazoans, as evidenced by the lethal effects of knocking out specific SR proteins in model organisms like Drosophila and Caenorhabditis elegans. However, not all SR proteins are essential for survival, and some may be dispensable in certain contexts.
Structural Organization of SR Proteins
SR proteins have a characteristic structural organization, consisting of RNA-binding domains at the N-terminus and an RS domain at the C-terminus. These domains are modular, meaning that the RNA-binding domains can be swapped between different SR proteins, and the RS domains can function even when attached to a different RNA-binding domain.
Other proteins related to SR proteins, known as SR-related proteins (SRrps), also play a role in pre-mRNA metabolism. These proteins include components of snRNPs, splicing regulators, coactivators, RNA helicases, and protein kinases.
The RNA-Binding Domain of SR Proteins
The RNA-binding specificity of SR proteins is crucial for their function in splicing. Several studies have investigated how SR proteins bind to pre-mRNA, helping to determine their role in splice site recognition and spliceosome assembly. Understanding these binding preferences is essential for fully grasping how SR proteins contribute to the accuracy and efficiency of splicing.